Improving RNA Assembly via Safety and Completeness in Flow Decompositions
Oct 25, 2022·
,
 ,
,
·
0 min read
,
,
·
0 min read
Shahbaz Khan
Milla Kortelainen
Manuel Cáceres
Lucia Williams
Alexandru I. Tomescu
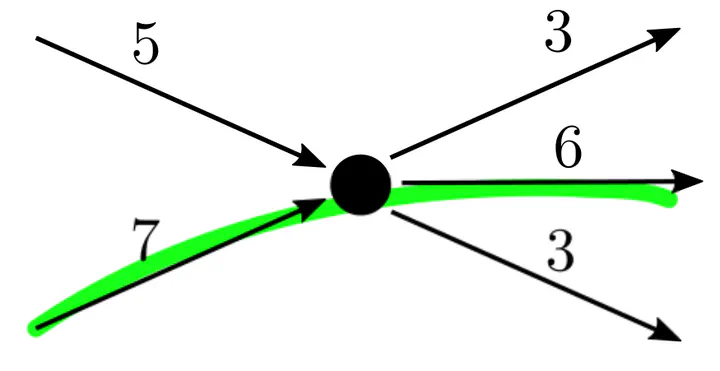 Safe flow subpath, which is not an extended unitig
Safe flow subpath, which is not an extended unitig
Abstract
Decomposing a network flow into weighted paths is a problem with numerous applications, ranging from networking, transportation planning to bioinformatics. In some applications we look for a decomposition that is optimal with respect to some property, such as number of paths used, robustness to edge deletion, or length of the longest path. However, in many bioinformatic applications, we seek a specific decomposition where the paths correspond to some underlying data that generated the flow. In these cases, no optimization criteria guarantees the identification of the correct decomposition. Therefore, we propose to instead report the safe paths, which are subpaths of at least one path in every flow decomposition.
In this work, we give the first local characterization of safe paths for flow decompositions in directed acyclic graphs (DAGs), leading to a practical algorithm for finding the complete set of safe paths. Additionally, we evaluate our algorithm on RNA transcript datasets against a trivial safe algorithm (extended unitigs), the recently proposed safe paths for path covers [TCBB 2021] and the popular heuristic greedy-width. One the one hand, we found that besides maintaining perfect precision, our safe and complete algorithm reports significantly higher coverage ($\approx 50\%$ more) as compared to the other safe algorithms. On the other hand, the greedy-width algorithm although reporting a better coverage, it also reports significantly lower precision on complex graphs (for genes expressing a large number of transcripts). Overall, our safe and complete algorithm outperforms (by $\approx 20\%$) greedy-width on a unified metric (F-Score) considering both coverage and precision when the evaluated dataset has a significant number of complex graphs. Moreover, it also has a superior time ($4-5\times$) and space performance ($1.2-2.2\times$), resulting in a better and more practical approach for bioinformatics applications of flow decomposition.
Type
Publication
In Journal of Computational Biology