Welcome
I am an HIIT Postdoctoral Fellow at Aalto University working with Professor Sándor Kisfaludi-Bak. My research interests are broadly in the area of efficient algorithms and data structures for problems on graphs and strings. I am particularly interested in understanding and overcoming intractable problems inside P, but in general I enjoy developing efficient solutions for intractable problems as well as understanding the limits of modern algorithmic techniques.
Interests
- Graph Algorithms
- String Algorithms
- Paramaterized Algorithms
- Algorithmic Bioinformatics
- Compressed Data Structures
- Approximation Algorithms
Education
-
PhD. in Computer Science, 2023
University of Helsinki
-
MSc. in Computer Science, 2019
University of Chile
-
Computer Engineering, 2019
University of Chile
-
BSc. in Computer Science, 2016
University of Chile
Featured Publications
Recent Publications
(2025).
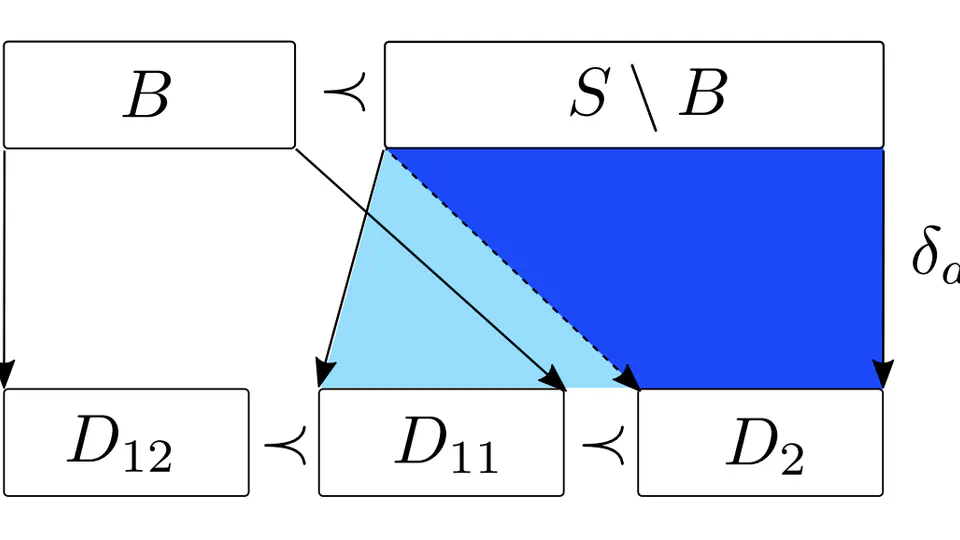
Identifying all snarls and superbubbles in linear-time, via a unified SPQR-tree framework.
In arXiv.
(2025).
Practical colinear chaining on sequences revisited.
In ISBRA 2025.
(2025).
Exploiting uniqueness: seed-chain-extend alignment on elastic founder graphs.
In ISMB 2025.
(2025).
Maximum Coverage k-Antichains and Chains: A Greedy Approach.
In arXiv.
(2024).
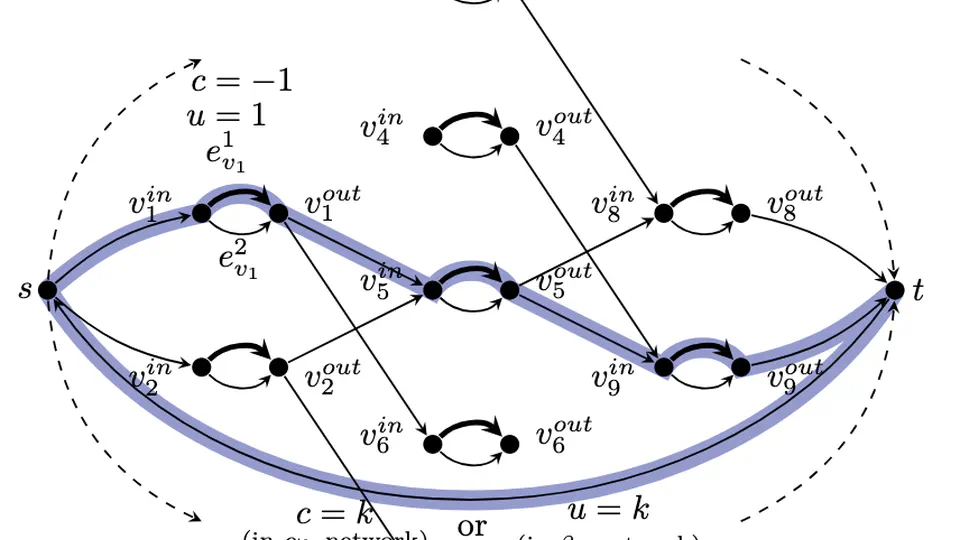
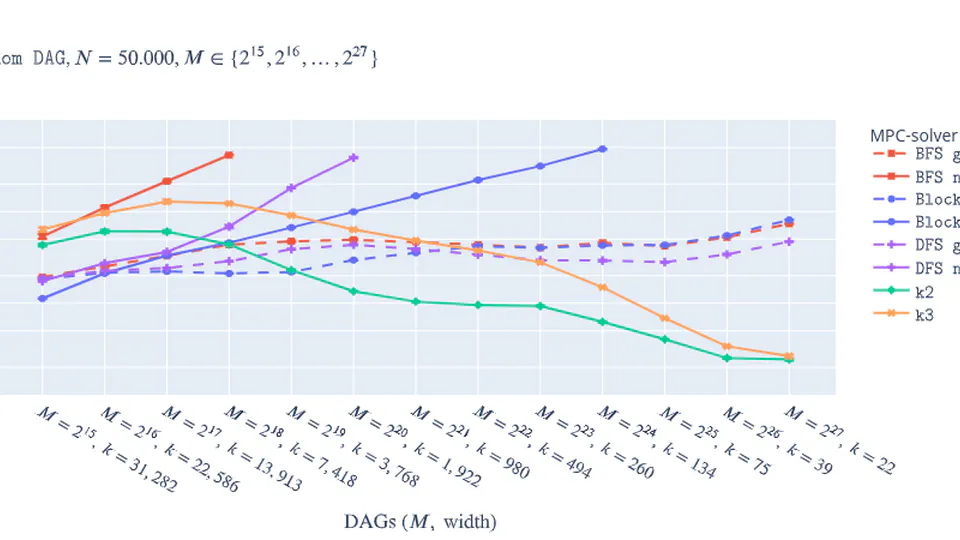
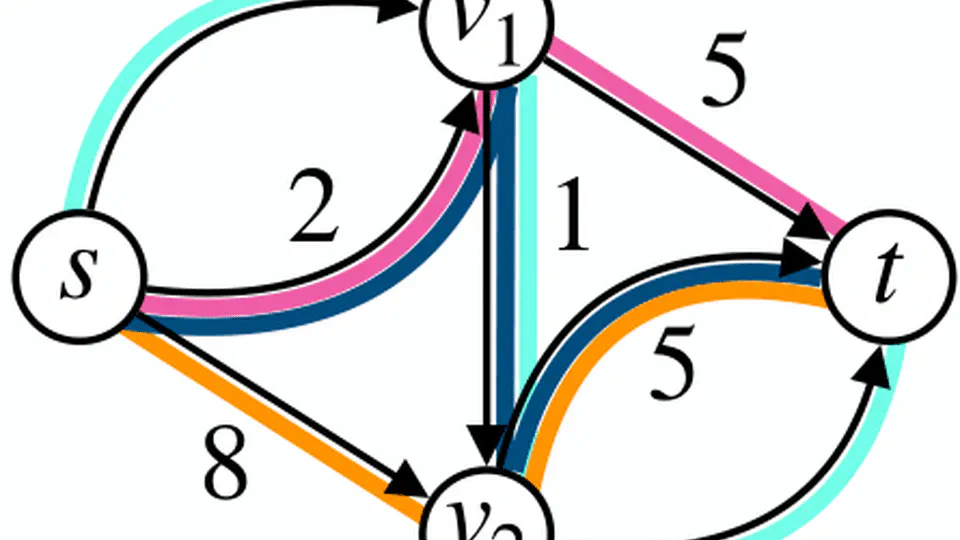
Practical Minimum Path Cover.
In SEA 2024.