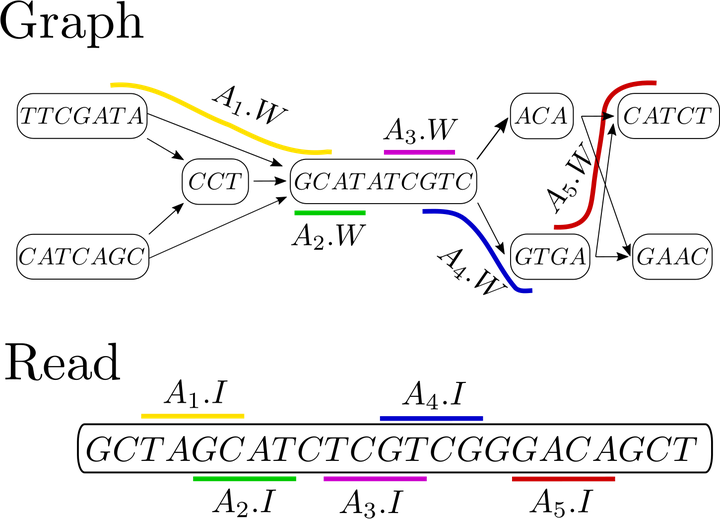
 Anchors in a string labeled acyclic graph
Anchors in a string labeled acyclic graph
Abstract
Aligning reads to a variation graph is a standard task in pangenomics, with downstream applications in e.g., improving variant calling. While the vg toolkit (Garrison et al., Nature Biotechnology, 2018) is a popular aligner of short reads, GraphAligner (Rautiainen and Marschall, Genome Biology, 2020) is the state-of-the-art aligner of erroneous long reads. GraphAligner works by finding candidate read occurrences based on individually extending the best seeds of the read in the variation graph. However, a more principled approach recognized in the community is to co-linearly chain multiple seeds. We present a new algorithm to co-linearly chain a set of seeds in a string labeled acyclic graph, together with the first efficient implementation of such a co-linear chaining algorithm into a new aligner of long reads to acyclic variation graphs, GraphChainer. Compared to GraphAligner, GraphChainer aligns 12% to 17% more reads, and 21% to 28% more total read length, on real PacBio reads from human chromosomes 1, 22 and the whole human pangenome. On both simulated and real data, GraphChainer aligns between 95% and 99% of all reads, and of total read length.We also show that minigraph (Li et al., Genome Biology, 2020) and minichain (Chandra and Jain, RECOMB, 2023) obtain an accuracy of less than 60% on this setting. GraphChainer is freely available at https://github.com/algbio/GraphChainer. The datasets and evaluation pipeline can be reached from the previous address.